
Immunoprecipitation Guide
Immunoprecipitation (IP) is a method of precipitating target protein out of solution by using an antibody that specifically binds to that particular protein. This process is powerful to isolate and concentrate a particular protein and/or protein complex from a sample containing many thousands of different proteins.
There are five types of IP:
Table 1. Five types of IP.
| Type | Target | Application | |
 | Individual protein immunoprecipitation (IP) | A known protein | Isolating particular protein |
 | Protein complex immunoprecipitation (Co-IP) | Intact protein complexes | Protein–protein interactions. |
 | Chromatin immunoprecipitation (ChIP) | DNA-binding proteins (including transcription factors and histones) | Protein–DNA interactions |
 | Ribonucleoprotein immunoprecipitation (RIP) | RNA-binding proteins (Ribonucleoproteins) | Protein–RNA interactions |
 | Tagged proteins immunoprecipitation | Engineer tags | All the above |
Individual protein immunoprecipitation (IP)
Individual protein immunoprecipitation is a procedure by using an antibody that is specific for a known protein to isolate that particular protein out of a solution containing many different proteins. It can also be used to “enrich” a protein population prior to Western Blotting.
Protein complex immunoprecipitation (Co-IP)
Co-immunoprecipitation is an extension of IP that is immunoprecipitation of intact protein complexes. By targeting a known member of the protein complex with an antibody, co-IP makes it possible to pull the entire protein complex out of solution and thereby identify unknown binding partners and the function of the target protein.
Chromatin immunoprecipitation (ChIP)
Given the fact that DNA-binding proteins (including transcription factors and histones) in living cells can be cross-linked to the DNA that they are binding, chromatin immunoprecipitation (ChIP) is used to immunoprecipitate the protein–DNA complex out of cellular lysates. This assay requires the knowledge of the interest protein and DNA, as a selective antibody targeting the interest protein and specific primers for the DNA must be provided. ChIP assay is a common method for transcriptional regulation monitoring.
Ribonucleoprotein immunoprecipitation (RIP)
Similar to ChIP, but rather than targeting protein-DNA complex in ChIP, RNP immunoprecipitation (RIP) is a technique to immunoprecipitate the protein-RNA complex using an antibody targeting the ribonucleoproteins of interest. RNA can be extracted from purified RNA-protein complexes and further analyzed by cDNA sequencing or RT-PCR.
Tagged-protein IP
As the basis of IP assay is targeting the interest protein by a selective antibody, the key limitation for IP approaches is the difficulty in generating an antibody that specifically recognize the target protein with little or no cross-reactivity. Due to the lack of an available antibody, many proteins cannot be immunoprecipitated. To circumvent this problem, proteins can be tagged on either the C- or N- terminal end with an epitope to which a high-affinity antibody is available. The most commonly used tags include FLAG, c-Myc, Hemagglutinin (HA), V5 and Green fluorescent protein (GFP).
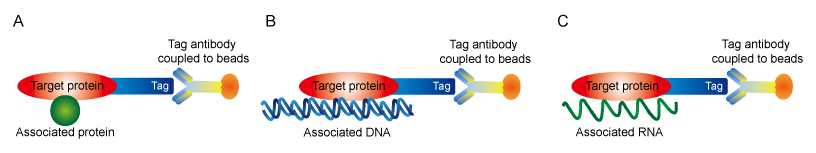 Figure 1. Tag strategies for co-IP (A), ChIP (B) and RNP-IP (C)
Figure 1. Tag strategies for co-IP (A), ChIP (B) and RNP-IP (C)
Basic procedure
The basic procedure of IP or Co-IP begins with the extraction of antigen from tissue samples or cells in an appropriate lysis buffer. Particularly for ChIP or RIP, the nucleic acid is cross-linked to protein by using formaldehyde or UV irradiation before cell lysis. The resulting lysate is incubated with an antibody, allowing the formation of an immune complex. Immobilized Protein A or Protein G linked to agarose or magnetic beads is simultaneously added to provide a solid phase support, which enables the precipitation or capture of those antibody-antigen complexes. The immunoprecipitated complexes are collected by centrifugation, and proteins are eluted for further analysis such as western blot and LC/MS. Nucleic acid analysis is achieved through PCR, RT-PCR, cDNA sequencing and microarray.
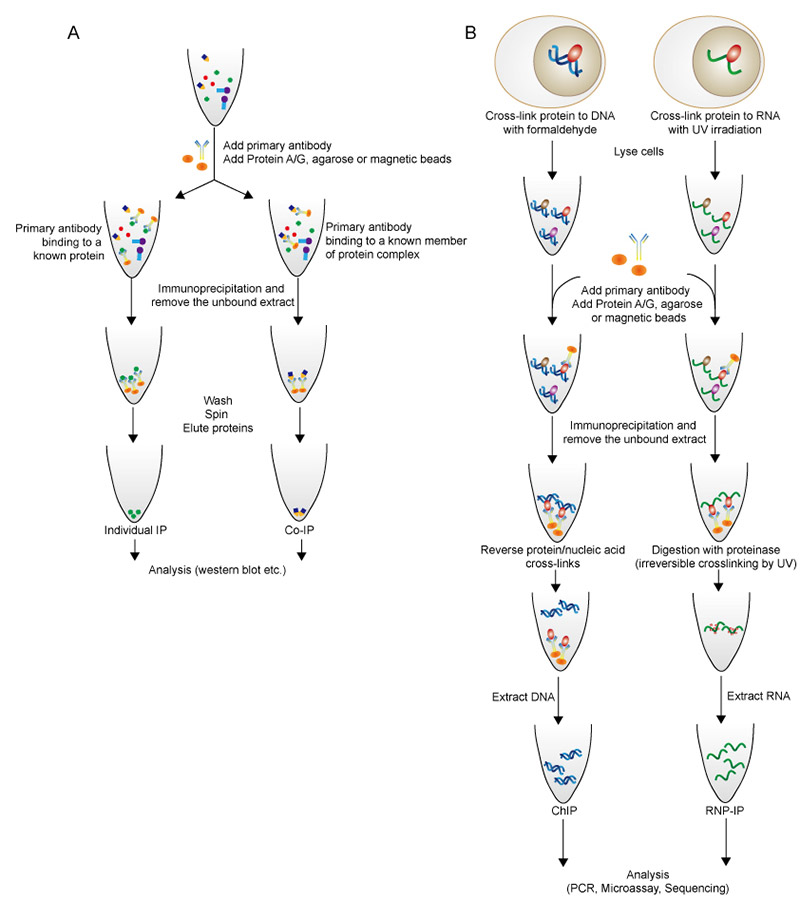 Figure 2. Flow diagram of individual IP and Co-IP (A), and ChIP and RNP-IP (B).
Figure 2. Flow diagram of individual IP and Co-IP (A), and ChIP and RNP-IP (B).
Protein A and Protein G
Protein A and Protein G provide an efficient binding of the antigen to the variable sites of the antibody molecule, as they are able to bind to the F region of most antibody classes from many species. Alternatively, the antibody can be covalently cross-linked to Protein A or G before sample addition. This strategy is achieved by using a common bifunctional cross-linker such as DSS, leading to the advantage of orienting the capture antibody via the Fc region for sterically unhindered antigen binding. However, cross-linker DSS may decrease the antigen biding efficiency under certain conditions. Notably, antibodies can be directly immobilized to an activated beaded support in lack of Protein A or G. This approach prevents the heavy and light chains of antibody from contaminating the eluted fraction. Generally, immobilized Protein A costs less than Protein G, while Protein G can bind to a broader range of classes of antibodies. Decisions on the application of Protein A or G are made according to the source and class of the antibody involved and the overall cost.
Agarose vs. magnetic bead
Agarose is most frequently used in protein purification because of the high protein binding capacity due to its high-porosity. Such porosity also provide a high surface area for antibody binding in IP assays. However, the high porosity can also become disadvantage when the quantity of antibody cannot meet the enormous binding capacity of the agarose. Magnetic beads are solid and usually spherical particles. The binding of antibody is limited to the surface of magnetic bead because the lack of porous core that in agarose. The application of magnetic beads in IP assays is a growing trend as magnetic beads display diverse advantages as below:
Table 2. Comparison between agarose and magnetic beads.
| What you may concern | Agarose | Magnetic bead |
| Size | 50 to 150 μm | 2-40 μm |
| Multiple appearance | No | Yes |
| Binding capacity | Great | Good |
| Diffusion | Slow | Fast |
| Required washing step | Extensive washing; Easy to lose sample | Washing steps are reduced; Less chance of proteolytic damage |
| Automation | No | Yes |
| Reproduciblity | Yes | Yes. Much better than agarose |
References
- Berggard, T., S. Linse, and P. James, Methods for the detection and analysis of protein-protein interactions. Proteomics, 2007. 7(16): p. 2833-42.
- Hassan, M.Q., et al., Ribonucleoprotein immunoprecipitation (RNP-IP): a direct in vivo analysis of microRNA-targets. J Cell Biochem, 2010. 110(4): p. 817-22.
- Khan, D.H. and J.R. Davie, Dual cross-linking ribonucleoprotein immunoprecipitation assay. Biochem Cell Biol, 2014. 92(4): p. 317-9.
- Nelson, J.D., O. Denisenko, and K. Bomsztyk, Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc, 2006. 1(1): p. 179-85.
- Phizicky, E.M. and S. Fields, Protein-protein interactions: methods for detection and analysis. Microbiol Rev, 1995. 59(1): p. 94-123.
- Kaboord B, Perr M. Isolation of proteins and protein complexes by immunoprecipitation. Methods Mol Biol, 2008. 424:349-64.






Leave Your Comment